In lab we set up a ISSR reaction on a large gel for the Lupinus arboreus. First, we directly added loading 1μL dye to the PCR tubes because we were not going to sequence the PCR reactions. Next, 5μL of each sample was loaded into the large 2% agarose gel. One person form the group loaded all the samples for all four members of the table. The gel was run at a low voltage as lecture proceed.
The second part of lab was spent editing additional sequences of the sequences that the class extracted this year. We downloaded EPIC_5525_New and built de novo assemblies which were trimmed at the ends. Heterozygous sites were edited as well. Then we exported the final consensus sequences and bulk renamed the sequences. Following this step the sequences form the previous week and the new sequences were grouped into a single folder. All the consensus sequences were selected and a Muscle alignment was created using default parameters. The alignment was edited once again to trim the edges. The file was closed and saved. The file was renamed 5525_nucleotide alignment_Lexly. A new folder was created and the new file was moved into it. The alignment was exported and uploaded to canvas for the concatenating process.
Following these steps, 3 different marker alignments were downloaded into the “Concatenated” folder that was created. Next, all four documents were selected and a concatenated sequence was generated. A new file was generated named “Concatenated alignment” which was set up for the process completed at home.
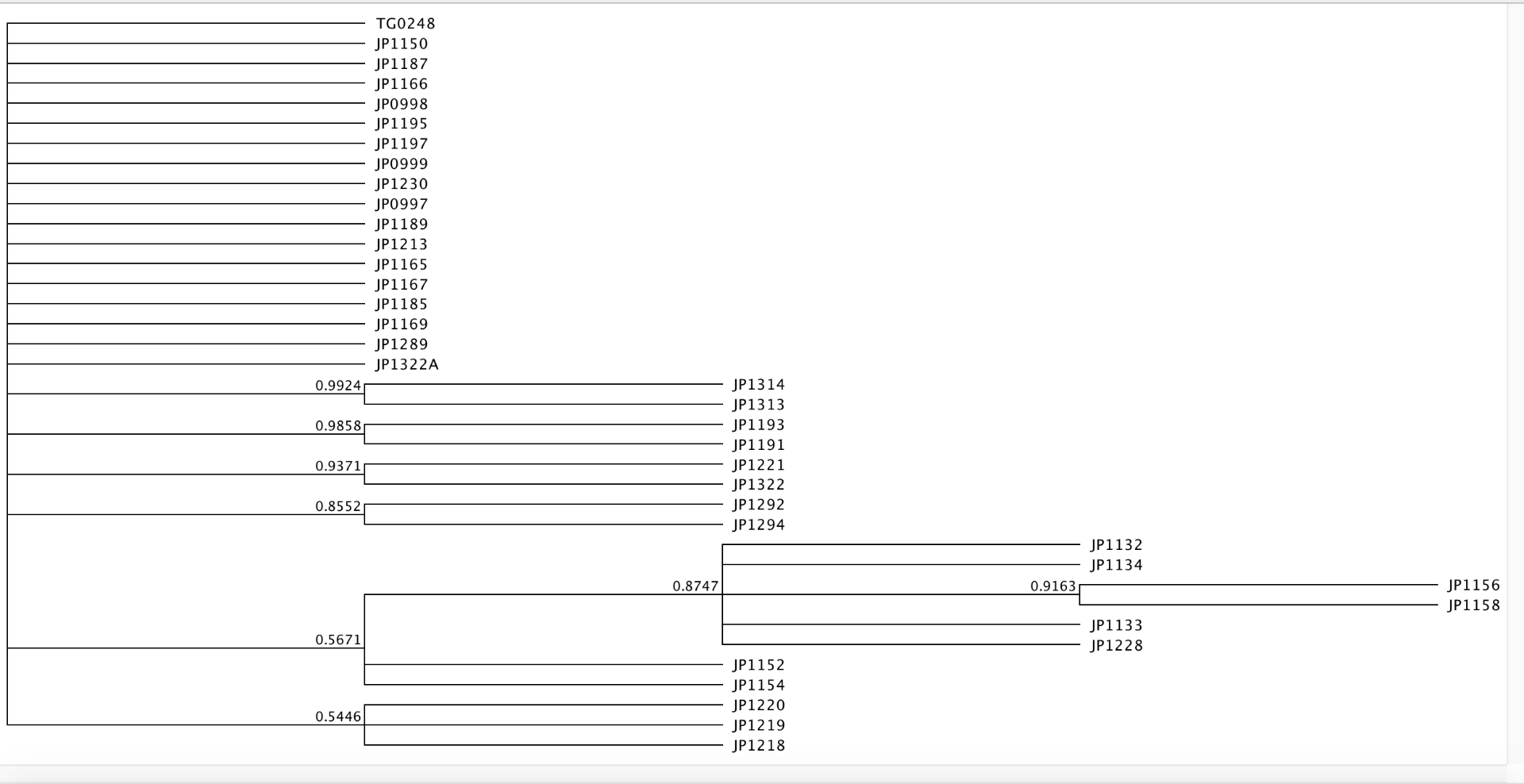
At home the exercised led us to determine the best model of molecular evolution by using the jModeltest. A phylogenetic tree was generated using MrBayes with 1.5 million generations, a burn-in of 100,000, and a subsample frequency of 500. TG0248 was set as the out-group. The model of evolution used was HKY.
Images: MrBayes Long Run
Analysis of Phylogenetic Tree:
Six clades presented clades with support values greater than 0.85. The individuals in each clade are most likely more similar due to being in the same population or geographically.



Leave a Reply