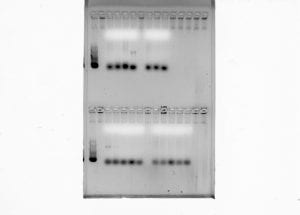
Subsequent to amplifying the genomic DNA (gDNA) collected from the fish samples, another gel electrophoresis was run, following the same steps as before, to see if specific bands were present. Unfortunately, some of the samples did not present the proper bands necessary to move onto the next procedure for PCR clean-up.
Another attempt was made by following the previous week’s steps for preparing the master mix and amplifying DNA via PCR. On the other hand, this would have been the protocol performed after obtaining the desired PCR results to clean up the amplified DNA and send for commercial sequencing:
- Prepare an ExoSap master mix containing water (10.59 μl), Sap 10x (1.25 μl), SAP (0.44 μl), Exo (0.22 μl), making sure that the reagents remain iced during preparation. The portions for each component should correspond with the numbers of reactions to be performed plus an additional amount covering for pipetting mistakes.
- Obtain a bucket of ice.
- Indicate and label the new PCR tubes to be used, then pipette 7.5 μl of each PCR product into the tubes.
- Pipette 12.5 μl of the ExoSap master mix into the newly labeled PCR tubes.
- Put the tubes in a thermocycler and start the ExoSap program, which should complete after approximately 45 minutes.
- Place the PCR tubes into a tube rack and put them in a freezer.