Allyson Luber
October 8, 2019
Lab 07: Phylogenetic Inference
In this lab we continued our work using Geneious program on the computer. Today we worked on our COI sequences a little more, including usage of another program which I’ll introduce later.
- Clean up alignment of COI sequences that includes fish DNA barcode and sequences that was downloaded from Geneious last week. Alignment should look neat and almost uniformal.
- Next, we used a different program called jModelTest2 to figure out the best model of molecular evolution for our sequences. JModelTest2 was downloaded from a website provided by our professor.
- Back to Geneious, we had to export our alignment in Phylip format, then back at jModel, export the file.
- In jModel, we worked with our alignment for awhile. First we computed likelihood scores. When they were done we chose the best model based on some optimality criterion. The two methods were Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC).
- For both methods, the analysis was done instantaneously where we were then guided to look at the results table where all the scores for the various models were shown.
Bayesian Inference
- Back in Geneious we selected a MrBayes analysis for a tree builder. Before we could do this we changed a couple things such as substitution model, rate variation, outgroup, and MCMC settings. Changing it will depend on what your professor prefers.
- Ran the analysis — it was quick!
- This time, we ran the analysis longer using 1,100,000 for the “chain length” and 100,000 for the burn-in. After the analysis was finished, the posterior output should be different from the first
Maximum Likelihood
- This last part of lab required using maximum likelihood to infer a phylogenetic tree of our aligned data set. In Geneious, download RAxML under tools –> plug-ins
- Next, under evolutionary model we chose “Rapid bootstrap with rapid hill climbing”
- Once RAxML was finished a new line was created and from here we had to build a consensus tree using that new line.
- The same procedure was repeated but instead of using RAxML we used PHYML (downloaded it first under plug-ins)
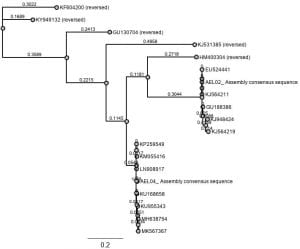
- Used HKY85 model of molecular evolution for this our the final Bayesian tree
Final tree result