Last week, we ran DD-RADSeq and Ligation on our Erythranthe guttata samples. This week, we began by running a test run of PCR to test for successful library construction.
- I began by labelling new PCR tubes to correspond to our samples #9-16 for this new PCR run.
- We created a Master Mix for the table, enough for 11 reactions, using the following recipe:
- NEB One-Taq 2x Master Mix: 8uL/rxn (88 uL total)
- Forward Primer (10mM): 0.4 uL/rxn (4.4 uL total)
- Reverse Primer (10mM): 0.4 uL/rxn (4.4 uL total)
- Pure H2O: 6.2uL/rxn (68.2 uL total)
3. We added 15 uL of the Master Mix and 1 uL of the library DNA template to each corresponding PCR tube.
4. We added these PCR tubes to the thermocycler BIORAD #1/2 –>DDRAD –>2_PCR1. This runs at 94 degrees Celsius for 2 min, then 20 cycles of (94 degrees C for 30 sec, 60 degrees C for 30 seconds, 68 degrees C for 45 sec). 4-10 degree C infinite hold.
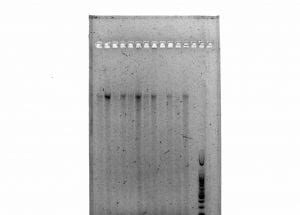
Gel Run
- After PCR was finished, we ran the PCR products in a 1.5% agarose gel (0.75 g agarose in 50 mL 1XTAE) with a 100 bp ladder at 130V for 40 minutes.
Final PCR
- To add Illumina flow cell annealing sequences, multiplexing indices, and sequencing primer annealing regions to all fragments and to increase concentrations of sequencing libraries, we perform a PCR amplification with a Fusion Polymerase kit.
- We prepared a Master Mix 2 for the table, enough for 11 reactions, following this recipe:
- Fusion DNA polymerase: 0.31 uL/rxn (3.4 uL total)
- 5X Fusion HF Buffer: 6.25 uL/rxn (68.8 uL total)
- 10uM Forward Primer (PCR1_X): 1.56 uL/rxn (17.2 uL total)
- 10uM Reverse Primer (PCR2_5): 1.56 uL/rxn (17.2 uL total)
- 10mM dNTPs: 0.63 uL/rxn (6.9 uL total)
- DMSO: 0.94 uL/rxn (10.3 uL total)
- Pure H2O: 10.8 uL/rxn (118.3 uL total)
4. For each reaction, we combined 3 uL of the library template DNA to a new, labeled PCR tube.
5. We then added 25 uL of the Master Mix 2 to each tube.
6. We vortexed, and then spun down in the centrifuge.
7. We ran PCR2 on thermocycler BIORAD #2. Specifically, this cycle ran as:
| Cycle Step | Cycles | Temp | Time |
| Initial Denaturation | 1 | 98 degrees C | 30 sec |
| Denaturation
Annealing Extension |
20 | 98 degrees C
65 degrees C 72 degrees C |
10 sec
30 sec 30 sec (b/c 1kb genome) |
| Final Extension | 1 | 72 degrees C | 5 min |
| Hold | 1 | 4 degrees C | infinite |
The next steps would be to run this PCR product on a gel and then select for bp size.