Lab 10 Entry
10/31/18 Wednesday. In this lab, I used Geneious to code heterozygotes and find polymorphism in the sequences.
Our EPIC marker failed, EPIC failure haha. So we used last year’s data, which were 5334 forward and reverse reads (25 reads of Mimulus cardinalis and outgroup of Mimulus lewisii).
First, I created forward and reverse sequences folder and dropped the reads into the file. I downloaded the reads from Canvas. Next, I selected all the sequences, both forward and reverse reads, and build an alignment using Muscle (default setting). Then, I looked at my nucleotide alignment and cut the ugly reads and looked for polymorphism and heterozygotes. I found many polymorphism and misreads, but about 10 true heterozygotes. Only 4-5 were true heterozygotes and polymorphic. I recoded the true heterozygotes with the appropriate ambiguity code from the IUPAC Ambiguity Code list I have been given in the beginning of the class. After I was done editing, I saved and applied the changes. Next, for each Forward and Reverse pair, I De Novo Assembly them and edited the assembly document so that it has good reads and correct Ambiguity Code. I repeated the same step from another pairs except the outgroup (TG0248) and forward read that did not have a pair (JP 1132). Then, I selected all the Assembly files and generate consensus sequence, and create sequence list. I then extract sequences from list and created a subfolder named 5334 Consensus Sequences. This will move all my Assembly documents into this subfolder. I then manually moved my outgroup reads and JP 1132 reads. Next, I selected them all and choose Edit, Batch Rename and click Remove button and put 33 to remove all the characters except the sequence name for the selected documents. However, I had to edit the name of the outgroup and JP 1132 by hand. After that, I selected all and created alignment using Muscle. I opened the alignment and did some trimming.
AT HOME – INFER A BAYESIAN TREE OF THIS SINGLE LOCUS ALIGNMENT:
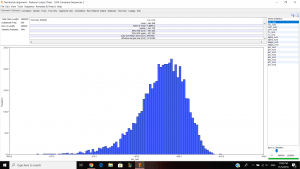
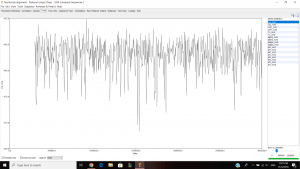
I inferred a Bayesian phylogenetic tree using 5334 nucleotide alignment and used the outgroup TG0248 and ran it for about 1hour and 30minutes. This is what I got after the run:
Left is the parameter estimates and right is the trace (good fuzzy caterpillar).
Tree 5334 consensus sequence-2krc97c
If screenshot the screen and paste it here, it came out blurry, so I just created a link to the Bayesian phylogenetic tree of 5334 consensus sequence.