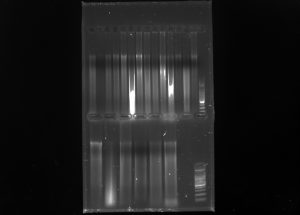
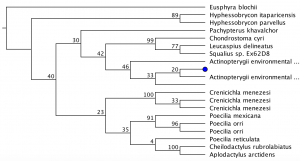
Inter simple sequence repeats (ISSR) uses microsatellite sequences as primers in PCR to generate multilocus markers that vary between organisms. Analysis and comparison of ISSR’s between organisms can reveal relatedness
Protocol:
Used ISSR markers (OMAR – (GAG)^4 RC) and psbA
On Samples (Left->Right)
GWC01 , CRA05 , PRO01 , PFR01 ,PRM01
20 tube Master mix for ISS (1 microliter template per tube)
ddH20 -> 250 microliters
10x buffer + Mg -> 60 microliters
BSA -> 20 microliters
dNTPs ->40 microliters
OMAR Primer -> 5 microliters
Taq -> 5 microliters
20 tube Master mix for psbA with following volumes in each tube
ddH20 -> 15.4 microliters
10x buffer + Mg -> 2 microliters
BSA -> 1 microliters
dNTPs ->.20 microliters
F Primer -> .2 microliters
R Primer -> .2 microliters
Taq -> .04 microliters
Placed on thermocycler