On Wednesday, November 8th, we continued with our project to analyze the genetic basis for polymorphism in Lupinus arboreus populations.
Gel Electrophoresis
In this gel electrophoresis project, we loaded PCR products into large agarose gels. These gels will allow us to compare many products at one time and more easily compare their differences in fragment length. As noted in last week’s procedure, using the ISSR markers should give us better population resolution for variation in L. arboreus.
Procedure:
- I pipetted 1 μL of loading dye in 3 PCR tubes with the Omar ISSR marker. I chose tubes CRA01, CRA02, and CRA03. I also pipetted 1 μL of loading dye in 3 PCR tubes with the 17898 ISSR marker. I chose tubes CRA03, CRA04, and CRA05.
- I gave my PCR tubes to Danielle and Prof. Paul, who pipetted their contents into the agarose gels. I also recorded which wells contained my samples.
- We ran the products out over the gel with gel electrophoresis for at least 90 minutes at 60 V.
Geneious Analysis of Lupinus ITS Sequences
We returned to using Geneious in order to build an alignment of the 41 L. arboreus individuals (and Lupinus chamissonis outgroup). We will use this ITS alignment for phylogenetic analysis of these individuals.
Procedure:
Creating Consensus Sequences from Forward-Reverse Reads
- I imported the Forward and Reverse L. arboreus ITS sequences into Geneious and aligned each Forward-Reverse pair into a De Novo Assembly.
- I then trimmed the regions of bad quality at the beginning and the end of the assembly. I also corrected any incorrect base calls within the sequence (from N to the appropriate base). I did this for each assembly generated in Step 1.
- I clicked on “Consensus”, “Extract”, and saved the document in order to create a consensus sequence for the first Forward-Reverse pair.
- I repeated Step 3 for each Forward-Reverse pair in the folder.
Building an alignment
- I built an alignment using Muscle with all the consensus sequences generated in the earlier procedure.
- I edited the alignment slightly by trimming the ends.
- I labeled this new alignment as “lupineITSNucleotidealignment”.
Exercises
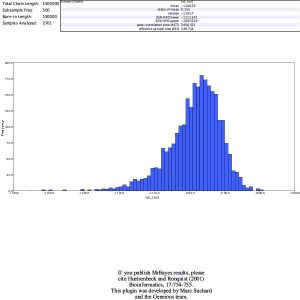
I built a phylogenetic tree with MrBayes and gained this result:

Posterior distribution and Trace:
My tree did not include clades with support values > 0.85.
In some cases, individuals from populations with purple flowers group together. For example, individuals from Pano–Near Diaz Ridge (PHO) and Pano2–Stinson Fire Road (PHT) grouped together. However, other individuals with purple flowers did not group together. This was true in the case of individuals from Limantour Beach–Point Reyes (PRL).
Based on the phylogenetic relationships of the Lupinus populations proposed by this tree, ITS does not provide enough resolution to distinguish populations phylogenetically. This is because many of the individuals were grouped together in one massive clade and it was not clear how they were related to each other phylogenetically.