On Wednesday, October 4th, we used Geneious to infer the phylogeny of the species of fishes gathered from sushi, other Actinopterygii species, and one Chondrichthyes species (used as an outgroup).
Procedure:
- First, I edited the sequences of my two samples (CMN1 and CMN3) in order to use their reverse complements in my alignment.
- I also edited the alignment, deleting the bases that preceded where my sequences began and ended beyond my sequences.
- I observed that within the first 20 columns of my alignment, two columns were polymorphic.
Choosing the best model of molecular evolution
- I downloaded JModelTest2 and exported my alignment into it.
- I used this program to compute the likelihood scores of possible phylogenetic trees.
- Next, I used two optimality criteria to determine which model was the best fit to propose phylogenetic trees. These criteria were the Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC).
- Both AIC and BIC chose the same model as the best, Model #79.
Bayesian interference
- I installed the MrBayes plugin.
- I chose the following parameters:
Substitution model: GTR
Rate variation: gamma
Outgroup: KT307481 (T. torpedo)
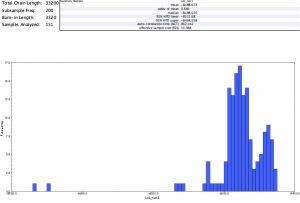
Chain length: 10,100
Subsampling freq.: 200
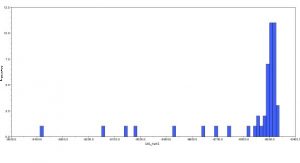
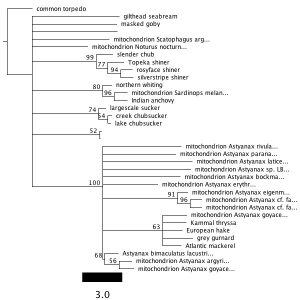
3. I ran the MrBayes analysis and received these results:
Inferred tree with posterior probability values:
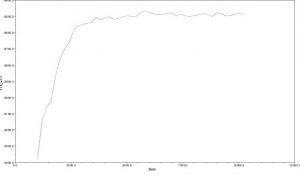
4. I performed the MrBayes analysis again, choosing a chain length of 1,100,000 and a burn-in value of 100,000. I received this result:
I did not recover the same clades in the two trees that resulted from MrBayes analysis. They also had different support values–the second analysis had much higher support values for each clade.
Maximum likelihood
- I installed the RAxML plugin.
- I chose the following parameters:
Evolutionary model: Rapid bootstrap with rapid hill climbing
100 BS Replicates
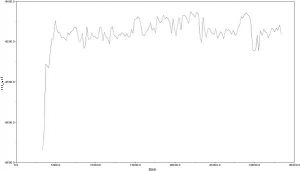
3. I built a consensus tree using the RAxML bootstrapping trees. I received this result:
I noticed that the clades with high support did match those from my Bayesian analysis!
4. I installed the PHYML plugin.
5. I ran a maximum likelihood analysis with bootstrapping, using the HKY85 model of molecular evolution. I received this result: