This week in lab we used our fish DNA sample results to use Geneious and analyze the genomic data. None of my samples fostered adequate results (I think this was due to exposure to soy sauce and having to stay in my backpack for more hours than originally planned). Because of this issue, I used a sample that was sequenced prior, called NS01, NS02, and NS03.
NS01 was labeled as yellow tail, NS01 was halibut, and NS03 was bluefin tuna. To start out the analysis I was personally worried because I tend to struggle with computers and new software. I made myself slow down and follow each step of the handed out protocol and found it to be easier than expected. I was surprised with the ability of the site to perform the BLAST so quickly and utilize the NCBI data base within the same forum and at such a fast rate.
I faced a few bumps in the road outside of not being able to use my original data. I accidentally deleted my first sequence assembly as well as the folders with my reverse and forward transcripts, but luckily after having practiced with the software I was able to quickly re-download everything and start from scratch.
Results:
NS01 served as yellowtail
Top result from the BLAST: Seriola quinqueradiata (Japanese amberjack-yellowtail)
Polymorphisms
- BP: 445 (A instead of G)
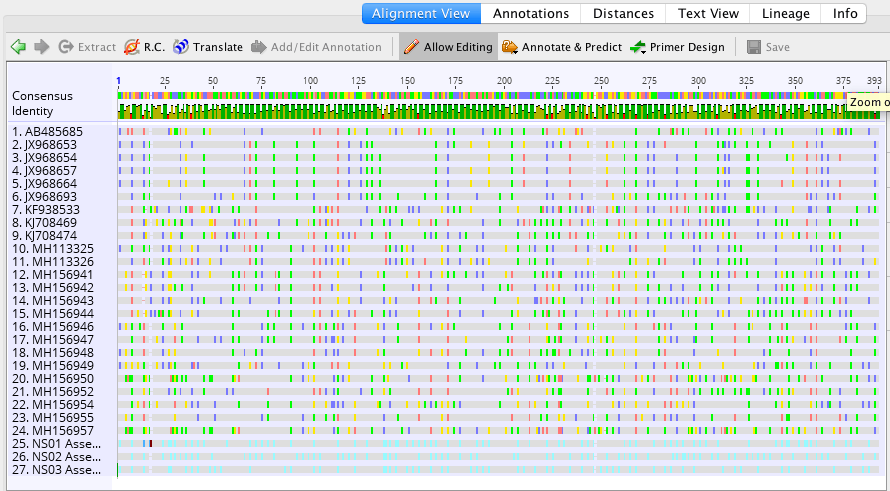
There was only one polymorphism after deleting all of the ambiguities (N) with a hit start and hit end in the high thousands. Therefore my first sample was served as the correct fish.
NS02 served as Halibut
Top result from the BLAST: Paralichthys californicus voucher – California halibut
Polymorphisms
The reverse of the genomic data NS02 carried greater than 20 polymorphisms throughout the entire comparison whereas the forward and all other vouchers had no polymorphisms despite a 100% match.
- BP: 403 (T instead of C)
2. BP: 413 (T instead of C)
3. BP: 418 (G instead of C)
4. BP: 421 (G instead of T)
5. BP: 428 (C instead of T)
6. BP: 435 (G instead of A)
7. BP: 438 (T instead of C)
8. BP: 442 (T instead of C)
9. BP: 443 (C instead of A)
10. BP: 458 (C instead of A)
NS03 served as Bluefin Tuna
Top result from the BLAST: Thunnus orientalis isolate – bluefin tuna
There were no polymorphisms present when comparing the matching sequences. With a 100% match and no polymorphisms it was surprising to see a slightly less than optimal hit start and end number below 1,000.









 Mt. Tamalpais (looking for Mimulus cardinals)
Mt. Tamalpais (looking for Mimulus cardinals) C
C