Oct. 10th, 2018 Lab
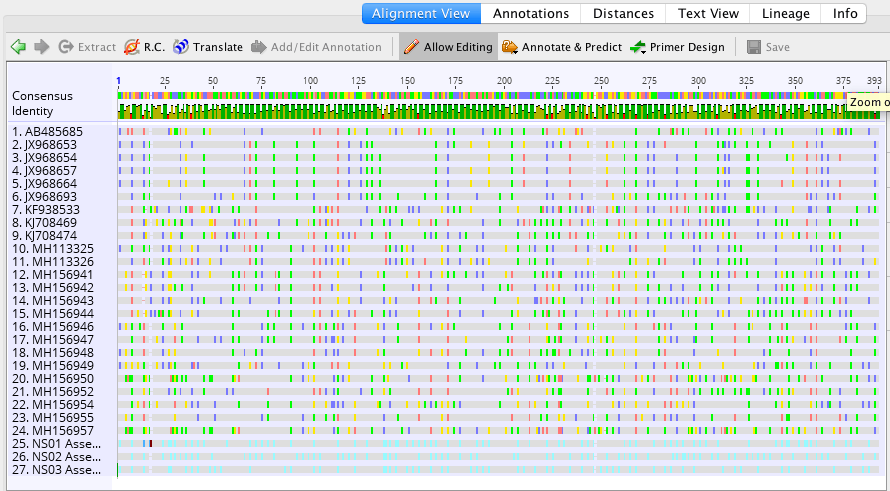
This week in lab after going over our lecture, we continued with our fish DNA samples to evaluate Phylogenetic Inferences of our data. I used the handed out protocol to par because doing so really helped me move through the software the week prior. Before coming to class I had already picked out around 25 different COI genes from a multitude of different Actinopterygii species. I also picked out 1 organism from the class chondrichthyes as an outgroup. I used Neotrygon kuhlii which is the bluespotted stingray. I had originally had picked another organism for this, but after aligning the sequences I realized that the genomes did not line up. Luckily, I was able to find the COI gene for the stingray and it lined up perfectly with my other sequences.
It took me awhile to troubleshoot finding the proper organisms through the NCBI nucleotide section in Genius because a bunch of random organisms would show up and if I got too specific in my search I got academic articles, not sequences.
Once my sequences were lined up I realized there were many polymorphic columns, at least in the first 20 columns. There were a total of 15 polymorphic columns in the first 20, but not all sets of 20 had this many. Others had anywhere from 5-1o columns that were polymorphic (if counting in sets of 20).
On the jModelTest my best model for AIC was JC 10975.525 . For my BIC it was JC 11182.16.
Initially I had a hard time figuring out how to get the plugins for the different tree models, but thanks to Dr. Paul I was able to figure it out:)
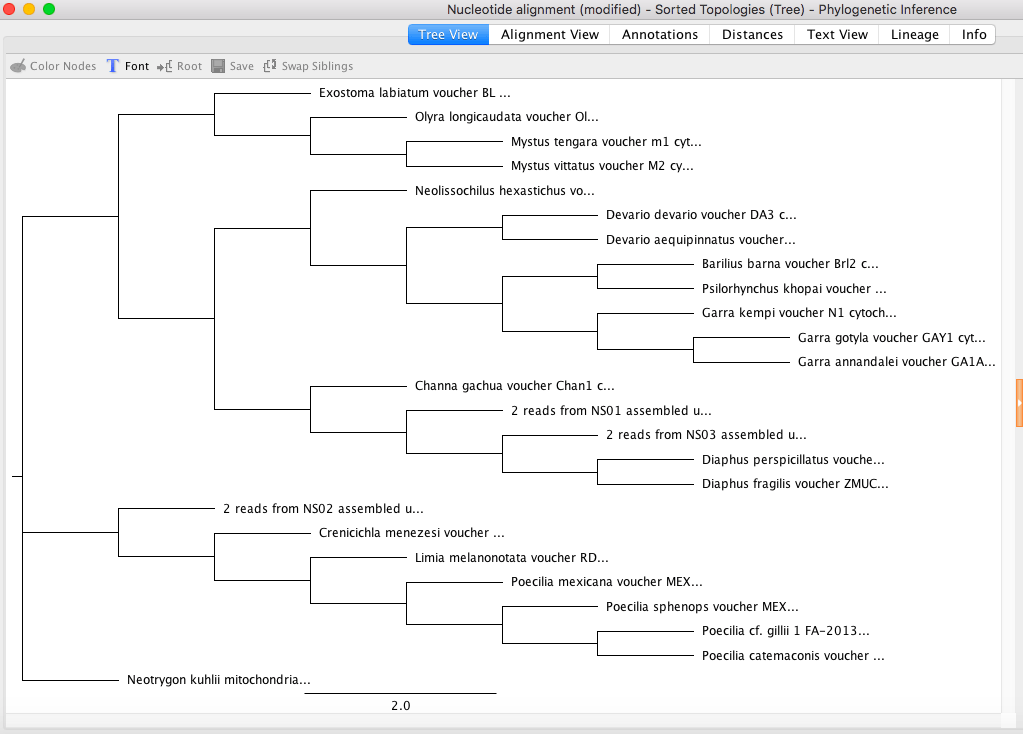
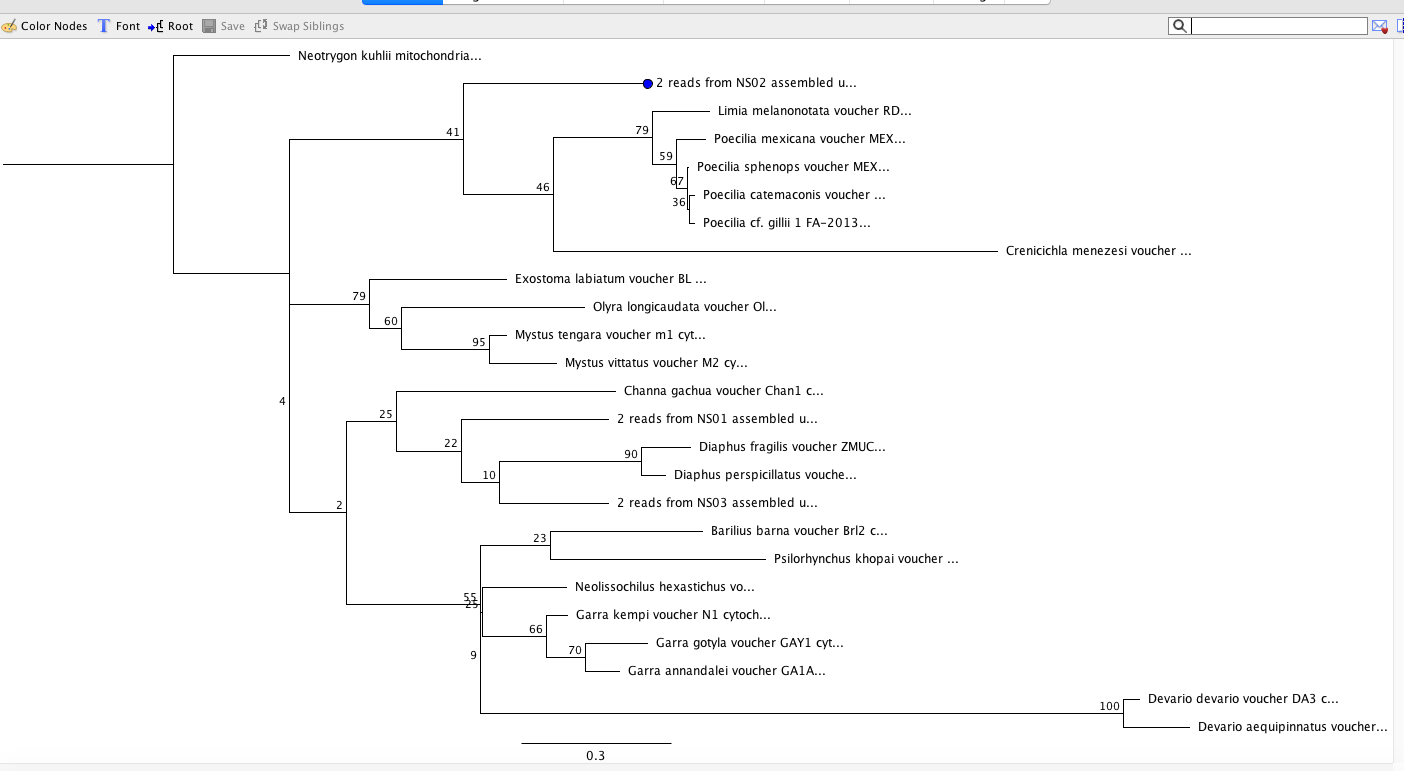
My maximum likelihood clades with the highest support also match my Bayesian analysis.
Issues: My folders kept being deleted so I had to redo this multiple times.
Organisms used for comparison:
Aligned Sequences:
AIC:
BIC:
Parameter estimates:
Trace:
Tree:
Maximum Likelihood/Consensus Tree: