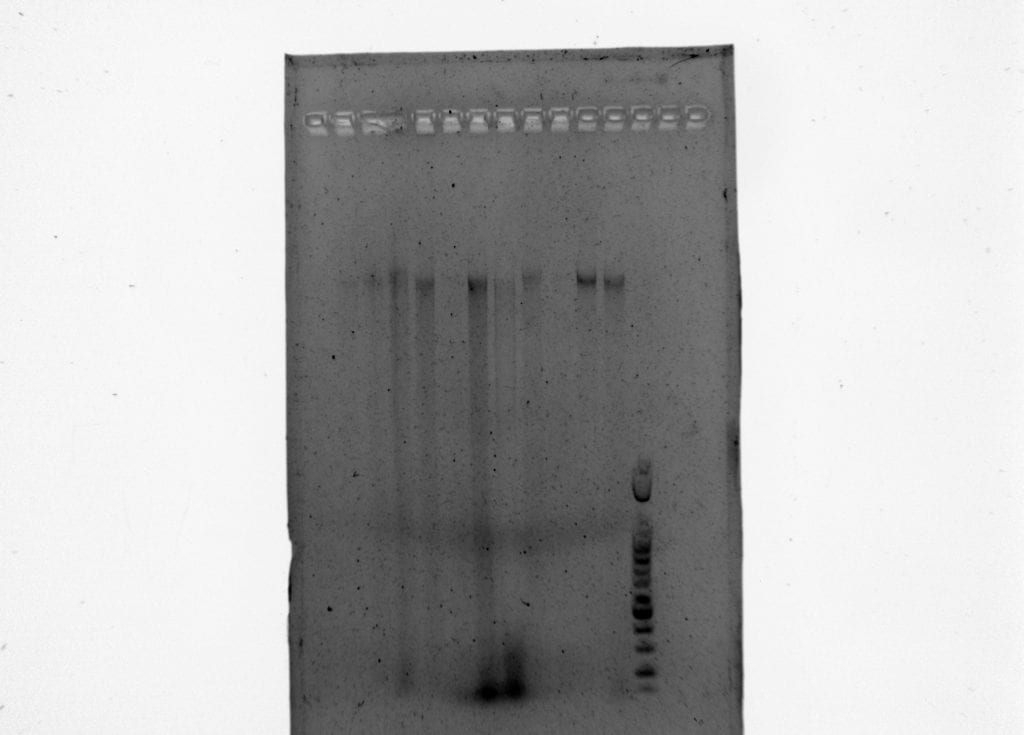
This lab we ran a test PCR to see if there was a successful library construction on our Mimulus guttatus samples using test PCR.
PCR I – Test Library Construction Success
NEB One-Taq 2x Master Mix – 80 uL
Forward Primer – 4 uL
Reverse Primer (PCR 026) – 4 uL
Pure H2O – 62UL
150 uL
Total reaction volume 16 uL
PCR1 ws used on BIORAD #1/2 at 94 degrees Celsius for 2 minutes, then 20 cycles of 94 degrees celsius for 30 seconds, 60 C fr 30 seconds, 68 C for 45 sec, and then at 4-10 C at infinite hold.
We then ran the products of PCR I for each sample on a 1.5% agarose gel with a 100 bp ladder at 130 V for 40 minutes.
PCR II – to generate final Illumina sequencing library
10 PCR reactions were set up in 25 uL volume.
Phusion DNA polymerase 3.1 uL
5x Phusion HF buffer 62.5 uL
Forward primer (PCR1_X) 15.6 uL
Reverse primer (2-7) 15.6 uL
DNTPS 6.3 uL
DMSO 9.4 uL
Pure H2O 107.5 uL
Master Mix Total 220 uL
Total reaction volume 25 uL
These were pipetted with individual samples into PCR tubes and then vortexed. PCR2 was run on BIORAD #2 for a number of cycles. Then 2 uL of the products of PCR2 were run on a 1% agrose gel with a 100 bp ladder.
Results of PCR 1 and PCR 2 are below.